Accelerating Lagrangian analyses of oceanic data: benchmarking typical workflows
Contents
Accelerating Lagrangian analyses of oceanic data: benchmarking typical workflows#
Purpose#
For data, Lagrangian refers to oceanic and atmosphere information acquired by observing platforms drifting with the flow they are embedded within, but also refers more broadly to the data originating from uncrewed platforms, vehicles, and animals that gather data along their unrestricted and often complex paths. Because such paths traverse both spatial and temporal dimensions, Lagrangian data often convolve spatial and temporal information that cannot always readily be organized in common data structures and stored in standard file formats with the help of common libraries and standards. As such, for both originators and users, Lagrangian data present challenges that the EarthCube CloudDrift project aims to overcome.
This notebook consists of systematic evaluations and comparisons of workflows for Lagrangian data, using as a basis the velocity and sea surface temperature datasets emanating from the drifting buoys of the Global Drifter Program (GDP). Specifically, we evaluate the data structure associated with common existing libraries in Python (xarray, pandas, and Awkward Array) in order to test their adequacies for performing three common Lagrangian tasks:
binning of a variable on an spatially-fixed grid (e.g. mean temperature map),
extracting data within given geographical and/or temporal windows (e.g. Gulf of Mexico),
analyses per trajectory (e.g. single statistics, spectral estimation by Fast Fourier Transform).
Since the CloudDrift project aims at accelerating the use of Lagrangian data for atmospheric, oceanic, and climate sciences, we hope that the users of this notebook will provide us with feedback on its ease of use and the intuitiveness of the proposed methods in order to guide the on-going development of the clouddrift Python package.
Technical contributions#
Description of some challenges arising from the analysis of large, heterogeneous Lagrangian datasets.
Description of some data formats for Lagrangian analysis with Python.
Comparison of performances of established and developing Python packages and libraries.
Methodology#
The notebook proceeds in three steps:
First, we download a subset of the hourly dataset of the GDP. Specifically, we access version 2.00 (beta) of the dataset that consists of a collection of 17,324 NetCDF files, one for each drifting buoy, available from a HHTPS (or FTP) server of the GDP. Alternative methods to download these data are described on the website of the GDP DAC at NOAA AOML and includes a newly-formed collection from the NOAA National Centers for Environmental Information with doi:10.25921/x46c-3620. We download a subset (which size can be scaled up or down) then proceed to aggregate the data from the individual files in one single file using a suggested format (the contiguous ragged array).
Second, we benchmark three libraries—xarray, Pandas, and Awkward Array—with typical Lagrangian workflow tasks such as the geographical binning of a variable, the extraction of the data for a given region, and operations performed per drifter trajectory.
Third, we discuss briefly future works for the upcoming clouddrift library, and seek to obtain feedback from the community to guide future development.
Results#
In terms of python packages, we find that Pandas is intuitive with a simple syntax but does not perform efficiently with a large dataset. The complete GDP hourly dataset is currently only ~15 GB, but as part of CloudDrift we also want to support larger Lagrangian datasets (>100 GB). On the other hand, xarray can interface with Dask to efficiently lazy-load large dataset but it requires custom adaptation to operate on a ragged array. In contrast, Awkward Array provides a novel approach by storing alongside the data an offset index in a manner that is transparent to the user, simplifying the analysis of non-uniform Lagrangian datasets. We find that it is also fast and can easily interface with Numba to further improve performances.
In terms of benchmark speed, each package show similar results for the geographical binning (test 1) and the operation per trajectory (test 3) benchmarks. For the extraction of a given region (test 2), xarray was found to be slower than both Pandas and Awkward Array. We note that speed and performance may not the deciding factor for all users and we believe that ease of use and simple intuitive syntax is as important for the end users. For this reason, we believe that the performance of Awkward Array and its ease of use make it a perfect framework to develop the CloudDrift library.
Funding#
Award1 = {"agency": "NSF EarthCube", "award_code": "2126413", "award_URL": "https://www.nsf.gov/awardsearch/showAward?AWD_ID=2126413"}
Award2 = {"agency": "Atlantic Oceanographic and Meteorological Laboratory", "award_code": "", "award_URL": "https://www.aoml.noaa.gov/global-drifter-program"}
Award3 = {"agency": "NOAA’s Global Ocean Monitoring and Observing Program", "award_code": "", "award_URL": "https://globalocean.noaa.gov"}
Keywords#
keywords=["lagrangian", "drifters", "data structures", "workflow tasks", "benchmarks"]
Citation#
P. Miron, S. Elipot, R. Lumpkin and B. Dano, Accelerating Lagrangian analysis of oceanic data: benchmarking typical workflows, 2022, 2022 EarthCube Annual Meeting, Accessed X/Y/2022 at https://github.com/Cloud-Drift/earthcube-meeting-2022
Acknowledgements#
The CloudDrift team would like to acknowledge Dr. Ryan Abernathey for suggesting to create benchmarks on typical Lagrangian workflow tasks and Dr. Jim Pivarski for assisting with Awkward array.
Setup#
Library import#
# Data manipulation
import numpy as np
import xarray as xr
import pandas as pd
import awkward as ak
from scipy import stats
# others
import time
from os.path import isfile, join
from datetime import datetime
import dask
import numba as nb
import functools
# data retrieval
import urllib.request
import concurrent.futures
import re
from tqdm import tqdm
# visualization
import matplotlib.pyplot as plt
import matplotlib.colors as colors
from mpl_toolkits.axes_grid1 import make_axes_locatable
from matplotlib.ticker import MaxNLocator
import cartopy.crs as ccrs
from cartopy.mpl.ticker import LongitudeFormatter, LatitudeFormatter
import cartopy.feature as cfeature
import cmocean
# we will see that the GDP DAC netcdf files contain non standard values that cause warnings
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
# default settings visualization
plt.rcParams.update({'font.size': 8,
'xtick.major.pad': 1,
'ytick.major.pad': 1})
# system information
import platform
import psutil
Local library import#
# To run this notebook, we need to load on a number of functions from the `preprocess.py` file found within this repository.
from preprocess import create_ragged_array, create_ak
Data Overview#
In the first step of this notebook, we present the current format of the Global Drifter Program (GDP) dataset, and show how to transform it into a single archival file in which each variable is stored in an ragged fashion.
The GDP produces two interpolated datasets of drifter position, velocity and sea surface temperature from more than 20,000 drifters that have been released since 1979. One dataset is at 6-hour resolution (Hansen and Poulain 1996) and the other one is at hourly resolution (Elipot et al. 2016). The files, one per drifter identified by its unique identification number (ID), are updated on a quarterly basis and are available via the HTTPS server of the GDP Data Assembly Center (DAC).
Here we use a subset of the hourly drifter dataset of the GDP by setting the variable subset_nb_drifters = 500. The suggested number is large enough to create an interesting dataset, yet without making the downloading cumbersome and the data processing too expensive. Feel free to scale down or up this value (from 1 to 17324), but beware that if you are running this notebook in a Binder there is a 2 GB memory limitation (running this Notebook with subset_nb_drifters = 500 requires close to 2 GB).
Note that you must execute Sec. 4 to create the archive (gdp_subset.nc) required for Sec. 5, 6, and 7. Once the ragged array is created, those sections can be executed in any order.
subset_nb_drifters = 500 # you can scale up/down this number (maximum value of 17324)
# output folder and official GDP https server
# Note: If you are running this notebook on a local computer and have already downloaded the individual NetCDF files
# independently of this notebook, you can move/copy these files to the folder destination shown below,
# or alternatively change the variable 'folder' to your folder with the data
folder = 'data/raw/'
input_url = 'https://www.aoml.noaa.gov/ftp/pub/phod/lumpkin/hourly/v2.00/netcdf/'
# load the complete list of drifter IDs from the AOML https
urlpath = urllib.request.urlopen(input_url)
string = urlpath.read().decode('utf-8')
pattern = re.compile('drifter_[0-9]*.nc')
filelist = pattern.findall(string)
list_id = np.unique([int(f.split('_')[-1][:-3]) for f in filelist])
# Here we "randomly" select a subset of ID numbers but produce reproducible results
# by actually setting the seed of the random generator
rng = np.random.RandomState(42) # reproducible results
subset_id = sorted(rng.choice(list_id, subset_nb_drifters, replace=False))
def fetch_netcdf(url, file):
'''
Download and save file from the given url (if not present)
'''
if not isfile(file):
req = urllib.request.urlretrieve(url, file)
# Typically it should take ~2 min for 500 drifters
print(f'Fetching the {subset_nb_drifters} requested netCDF files (as a reference ~2min for 500 files).')
with concurrent.futures.ThreadPoolExecutor() as exector:
# create list of urls and paths
urls = []
files = []
for i in subset_id:
file = f'drifter_{i}.nc'
urls.append(join(input_url, file))
files.append(join(folder, file))
# parallel retrieving of individual netCDF files
list(tqdm(exector.map(fetch_netcdf, urls, files), total=len(files)))
Fetching the 500 requested netCDF files (as a reference ~2min for 500 files).
100%|████████████████████████████████████████████████████████████| 500/500 [00:00<00:00, 319104.08it/s]
Individual NetCDFs#
Dimensions#
Each individual NetCDF file contains two main dimensions for its variables:
['traj']of length 1, since there is one trajectory per file (e.g.ID,deploy_date,deploy_lon, etc.)['obs']: of length N, with N the number of observations along the trajectory (lon,lat,ve,vn,sst, etc.)
Variables#
In each file, there are 20 numerical variables representing estimated quantities with dimensions ['traj', 'obs'] (where traj = 1 since there is only one trajectory per file). There are 11 numerical variables with dimension ['traj'] that contain metadata unique to each drifter and file (e.g. Deployment date and time). In addition, there are 2 variables with one of their dimensions being ['traj'] that also contains non-numerical metadata (e.g. Buoy type). Further metadata pertaining to an individual drifter and to the whole dataset are contained in various global attributes of the NetCDF file.
To have a “peek” at the structure of the NetCDF files (dimensions, variables, and attributes), we load the first file in our list of files as an xarray dataset object using the open_datasetfunction. xarray offers a pleasant html visualization of a dataset (expand the view by clicking on the black arrows to the left).
ds = xr.open_dataset(files[0], decode_times=False)
ds
<xarray.Dataset>
Dimensions: (traj: 1, obs: 1095)
Dimensions without coordinates: traj, obs
Data variables: (12/33)
ID (traj) |S10 b'2592'
rowsize (traj) int32 1095
WMO (traj) float64 4.401e+06
expno (traj) float64 9.046e+03
deploy_date (traj) float32 9.887e+08
deploy_lat (traj) float64 47.43
... ...
err_sst (traj, obs) float64 0.074 0.077 0.086 ... 0.042 -1e+34
err_sst1 (traj, obs) float64 0.224 0.158 0.113 ... 0.107 -1e+34
err_sst2 (traj, obs) float64 0.236 0.185 0.142 ... 0.087 0.1 -1e+34
flg_sst (traj, obs) float64 5.0 5.0 5.0 5.0 ... 5.0 5.0 5.0 0.0
flg_sst1 (traj, obs) float64 5.0 5.0 5.0 5.0 ... 5.0 5.0 5.0 0.0
flg_sst2 (traj, obs) float64 4.0 4.0 4.0 4.0 ... 4.0 4.0 2.0 0.0
Attributes: (12/72)
title: Global Drifter Program hourly drifting buoy c...
id: Global Drifter Program ID 2592
location_type: Argos
wmo_platform_code: 4400509
ncei_template_version: NCEI_NetCDF_Trajectory_Template_v2
cdm_data_type: Trajectory
... ...
DrogueCenterDepth: 15 m
DrogueDetectSensor: submergence
acknowledgement: Elipot et al. (2016), Elipot et al. (2021) to...
history: Version 2.00. Metadata from dirall.dat and d...
interpolation_method:
imei: Contiguous Ragged Array#
In the GDP dataset, the number of observations varies from len(['obs'])=13 to len(['obs'])=66417. As such, it seems inefficient to create bidimensional datastructure ['traj', 'obs'], commonly used by Lagrangian numerical simulation tools such as Ocean Parcels and OpenDrift that tend to generate trajectories of equal or similar lengths.
Here, we propose to combine the data from the individual netCDFs files into a contiguous ragged array eventually written in a single NetCDF file in order to simplify data distribution, decrease metadata redundancies, and efficiently store a Lagrangian data collection of uneven lengths. The aggregation process (conducted with the create_ragged_array function found in the module preprocess.py) also converts to variables some of the metadata originally stored as attributes in the individual NetCDFs. The final structure contains 21 variables with dimension ['obs'] and 38 variables with dimension ['traj'].
create_ragged_array(files).to_xarray().to_netcdf(join('data', 'gdp_subset.nc'))
100%|████████████████████████████████████████████████████████████████| 500/500 [00:08<00:00, 60.40it/s]
# using the previously downloaded files
ds = xr.open_dataset('data/gdp_subset.nc')
ds
<xarray.Dataset>
Dimensions: (traj: 500, obs: 4786301)
Coordinates:
ID (traj) int64 2592 6428 13566 ... 68246720 68248530
lon (obs) float32 ...
lat (obs) float32 ...
time (obs) datetime64[ns] ...
ids (obs) int64 ...
Dimensions without coordinates: traj, obs
Data variables: (12/54)
rowsize (traj) int64 1095 19132 6631 17906 ... 643 1843 1176
location_type (traj) bool False False False ... True True True
WMO (traj) int32 4400509 1600536 ... 4601712 4601740
expno (traj) int32 9046 9435 7325 ... 21312 21312 21312
deploy_date (traj) datetime64[ns] 2001-05-01 ... 1970-01-01
deploy_lon (traj) float32 -52.17 71.24 -97.16 ... -151.0 -143.4
... ...
err_sst (obs) float32 ...
err_sst1 (obs) float32 ...
err_sst2 (obs) float32 ...
flg_sst (obs) int8 ...
flg_sst1 (obs) int8 ...
flg_sst2 (obs) int8 ...
Attributes: (12/15)
title: Global Drifter Program hourly drifting buoy collection
history: Version 2.00. Metadata from dirall.dat and deplog.dat
Conventions: CF-1.6
date_created: 2022-05-20T14:48:40.763710
publisher_name: GDP Drifter DAC
publisher_email: aoml.dftr@noaa.gov
... ...
metadata_link: https://www.aoml.noaa.gov/phod/dac/dirall.html
contributor_name: NOAA Global Drifter Program
contributor_role: Data Acquisition Center
institution: NOAA Atlantic Oceanographic and Meteorological Laboratory
acknowledgement: Elipot et al. (2022) to be submitted. Elipot et al. (2...
summary: Global Drifter Program hourly dataIn this xarray.Dataset object, the data from the five hundred trajectories (here ordered by ID number by choice) are stored one after the other along the obs dimension. To be able to track the sizes of each consecutive trajectory, we have created a new array variable in this dataset called rowsize which contains all trajectory lengths.
ds.rowsize
<xarray.DataArray 'rowsize' (traj: 500)>
array([ 1095, 19132, 6631, ..., 643, 1843, 1176])
Coordinates:
ID (traj) int64 2592 6428 13566 17927 ... 68244730 68246720 68248530
Dimensions without coordinates: traj
Attributes:
long_name: Number of observations per trajectory
sample_dimension: obs
units: -In summary, we find that the structure of such a contiguous ragged array is perhaps not as straightforward as a two-dimensional array, but nevertheless it has several advantages:
Only one file is needed to hold the complete dataset and less storage space is needed than for 2D matrix
The extra
rowsizevariable is a small addition to the original data that will be useful to access data per trajectory (see next section)It should be efficient to perform reduce operation (e.g.
mean,std, etc.) on the full dataset since the data is stored in a continuous array.
del ds
In the second and next step of the notebook, we benchmark different data science Python libraries. For this, the following sections present three typical Lagrangian tasks which are conducted successively using xarray, Pandas, and finally Awkward Arrays.
Xarray#
path_gdp = 'data/gdp_subset.nc'
ds = xr.open_dataset(path_gdp, chunks={}) # chunks={} to automatically select chunks size
The xarray package provides a nice html interface that allows us to quickly scroll to the variables of the xr.Dataset, look at its attributes, and get information about the underlying data structure. Xarray also supports reading the data in chunks which are pieces of the underlying data, represented as many NumPy (or NumPy-like) arrays. The size of the chunks is critical to optimize advanced algorithms. Using the default settings (with chunks={}), we can see that there is only one chunk per dimension, since the length of one of the variable can easily fit in memory (click on the disk symbol to the right to expand the information and visually see the chunk size).
ds
<xarray.Dataset>
Dimensions: (traj: 500, obs: 4786301)
Coordinates:
ID (traj) int64 dask.array<chunksize=(500,), meta=np.ndarray>
lon (obs) float32 dask.array<chunksize=(4786301,), meta=np.ndarray>
lat (obs) float32 dask.array<chunksize=(4786301,), meta=np.ndarray>
time (obs) datetime64[ns] dask.array<chunksize=(4786301,), meta=np.ndarray>
ids (obs) int64 dask.array<chunksize=(4786301,), meta=np.ndarray>
Dimensions without coordinates: traj, obs
Data variables: (12/54)
rowsize (traj) int64 dask.array<chunksize=(500,), meta=np.ndarray>
location_type (traj) bool dask.array<chunksize=(500,), meta=np.ndarray>
WMO (traj) int32 dask.array<chunksize=(500,), meta=np.ndarray>
expno (traj) int32 dask.array<chunksize=(500,), meta=np.ndarray>
deploy_date (traj) datetime64[ns] dask.array<chunksize=(500,), meta=np.ndarray>
deploy_lon (traj) float32 dask.array<chunksize=(500,), meta=np.ndarray>
... ...
err_sst (obs) float32 dask.array<chunksize=(4786301,), meta=np.ndarray>
err_sst1 (obs) float32 dask.array<chunksize=(4786301,), meta=np.ndarray>
err_sst2 (obs) float32 dask.array<chunksize=(4786301,), meta=np.ndarray>
flg_sst (obs) int8 dask.array<chunksize=(4786301,), meta=np.ndarray>
flg_sst1 (obs) int8 dask.array<chunksize=(4786301,), meta=np.ndarray>
flg_sst2 (obs) int8 dask.array<chunksize=(4786301,), meta=np.ndarray>
Attributes: (12/15)
title: Global Drifter Program hourly drifting buoy collection
history: Version 2.00. Metadata from dirall.dat and deplog.dat
Conventions: CF-1.6
date_created: 2022-05-20T14:48:40.763710
publisher_name: GDP Drifter DAC
publisher_email: aoml.dftr@noaa.gov
... ...
metadata_link: https://www.aoml.noaa.gov/phod/dac/dirall.html
contributor_name: NOAA Global Drifter Program
contributor_role: Data Acquisition Center
institution: NOAA Atlantic Oceanographic and Meteorological Laboratory
acknowledgement: Elipot et al. (2022) to be submitted. Elipot et al. (2...
summary: Global Drifter Program hourly dataXarray test 1: Geographical binning of any variable#
The first benchmark test implemented here is to compute statistics per geographical bins. Here we choose to compute the mean of the zonal velocity (ve) from all drifter estimates. From the previous cell, we can see that the size of the chunks were automatically set to the size of obs, leading to one chunk per variable. For this type of bin operation, one chunk is convenient because the complete zonal velocity ragged array variable will be loaded in memory in one operation. In order to perform operations per trajectory (shown below), we will instead set the size of the chunks to be the length of each trajectory, which here would have required subset_nb_drifters operation to load the same data.
Note: chunks should always align with the computation to optimize performance (more details in section 5.3.2).
In order to keep track of computing times of each test, we create a variable benchmark_times.
benchmark_times = np.zeros((3,3)) # 3 benchmark tests for the 3 different processing libraries
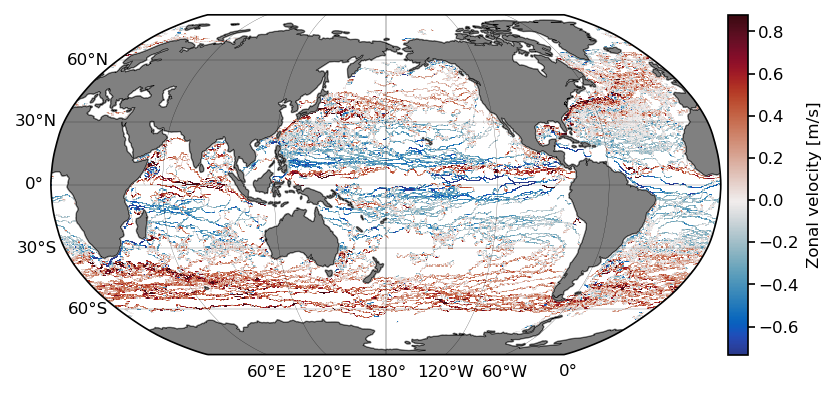
We calculate the mean zonal velocity using the binned_statistic_2d function from the stats module of the scipy package:
t0 = time.time()
lon = np.linspace(-180, 180, 360*2)
lat = np.linspace(-90, 90, 180*2)
ret = stats.binned_statistic_2d(ds.lon,
ds.lat,
ds.ve,
statistic=np.nanmean,
bins=[lon, lat])
benchmark_times[0,0] = time.time() - t0
We now plot the results:
x_c = np.convolve(lon, [0.5, 0.5], mode='valid')
y_c = np.convolve(lat, [0.5, 0.5], mode='valid')
# get 1st and 99th percentiles of values to plot to get a useful range for the colorscale
v1,v2 = np.nanpercentile(ret.statistic.T,[1,99])
fig = plt.figure(dpi=150)
ax = fig.add_subplot(1,1,1,projection=ccrs.Robinson(central_longitude=-180))
cmap = cmocean.tools.crop(cmocean.cm.balance, vmin=v1, vmax=v2, pivot=0)
pcm = ax.pcolormesh(x_c, y_c,
ret.statistic.T,
cmap=cmap,
transform=ccrs.PlateCarree(),
vmin=v1, vmax=v2)
# gridlines and labels
gl = ax.gridlines(color='k', linewidth=0.1, linestyle='-',
xlocs=np.arange(-180, 181, 60), ylocs=np.arange(-90, 91, 30),
draw_labels=True)
gl.top_labels = False
gl.right_labels = False
# add land and coastline
ax.add_feature(cfeature.LAND, facecolor='grey', zorder=1)
ax.add_feature(cfeature.COASTLINE, linewidth=0.25, zorder=1)
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", size="3%", pad=0.05, axes_class=plt.Axes)
cb = fig.colorbar(pcm, cax=cax);
cb.ax.set_ylabel('Zonal velocity [m/s]');
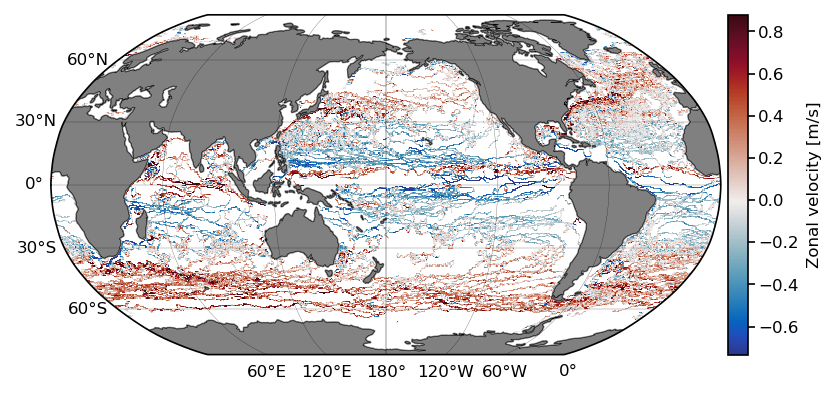
Xarray test 2: Extract a given region#
Extracting a subregion of the dataset is also a very common operation. xarray provides efficient way to apply mask to the data. One important thing to note is that since we have two dimensions, a mask is applied on the obs as well as on the traj dimensions. We first define a function to do the data extraction:
def retrieve_region(ds, lon: list = None, lat: list = None, time: list = None) -> xr.Dataset:
'''Subset the dataset for a region in space and time
Args:
ds: xarray Dataset
lon: longitude slice of the subregion
lat: latitude slice of the subregion
time: tiem slice of the subregion
Returns:
ds_subset: Dataset of the subregion
'''
# define the mask for the 'obs' dimension
mask = np.ones(ds.dims['obs'], dtype='bool')
if lon:
mask &= (ds.coords['lon'] >= lon[0]).values
mask &= (ds.coords['lon'] <= lon[1]).values
if lat:
mask &= (ds.coords['lat'] >= lat[0]).values
mask &= (ds.coords['lat'] <= lat[1]).values
if time:
mask &= (ds.coords['time'] >= np.datetime64(time[0])).values
mask &= (ds.coords['time'] <= np.datetime64(time[1])).values
# define the mask for the 'traj' dimension using the ID numbers from the masked observation
mask_id = np.in1d(ds.ID, np.unique(ds.ids[mask]))
ds_subset = ds.isel(obs=np.where(mask)[0], traj=np.where(mask_id)[0])
return ds_subset.compute()
Next we use this function to extract the data from the Gulf of Mexico for years 2000 to 2020:
t0 = time.time()
# we need to record the time for benchmarking in the end
lon = [-98, -78]
lat = [18, 31]
day0 = datetime(2000,1,1).strftime('%Y-%m-%d')
day1 = datetime(2020,12,31).strftime('%Y-%m-%d')
days = [day0, day1]
ds_subset = retrieve_region(ds, lon, lat, days)
benchmark_times[0,1] = time.time() - t0
For visualization of the results, the sea surface temperature estimates along trajectories from the extracted subregion are plotted in the next cell.
fig = plt.figure(dpi=150)
ax = fig.add_subplot(1,1,1,projection=ccrs.PlateCarree())
# temperature data are in Kelvin so we convert to degree Celsius
pcm = ax.scatter(ds_subset.lon, ds_subset.lat,
s=0.05, c=ds_subset.sst-273.15, transform=ccrs.PlateCarree(),
cmap=cmocean.cm.thermal, vmin=-2, vmax=50)
ax.add_feature(cfeature.LAND, facecolor='grey', zorder=1)
ax.add_feature(cfeature.COASTLINE, linewidth=0.25, zorder=1)
ax.set_xticks(np.arange(-95, -79, 5), crs=ccrs.PlateCarree())
ax.set_yticks([20, 25, 30], crs=ccrs.PlateCarree())
ax.xaxis.set_major_formatter(LongitudeFormatter())
ax.yaxis.set_major_formatter(LatitudeFormatter())
divider = make_axes_locatable(ax)
cax = divider.append_axes('right', size='3%', pad=0.02, axes_class=plt.Axes)
cb = fig.colorbar(pcm, cax=cax)
cb.set_label('Sea Surface Temperature [˚C]')
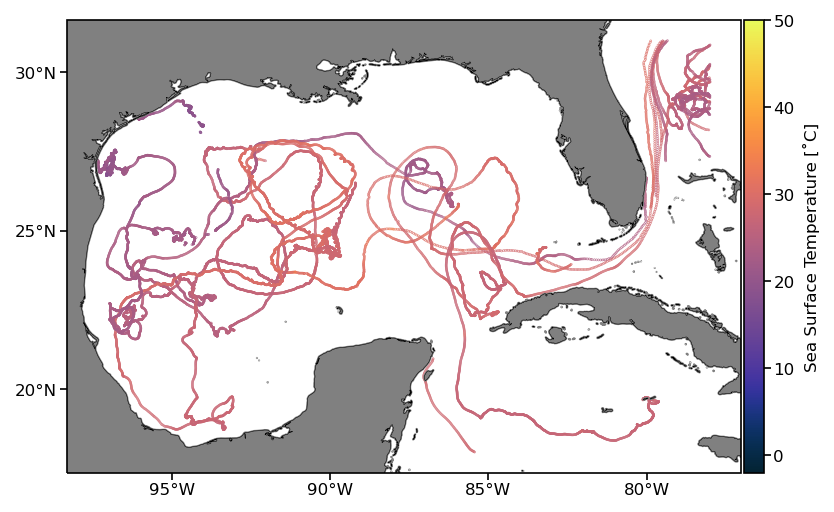
Xarray test 3: Operations per trajectory#
Single statistic per trajectory#
The next type of common operation with Lagrangian data is to compute statistics, or perform time series analysis, per trajectory. In order to do this with an xarray dataset, we need to create first a variable that indexes the beginning of each trajectory, i.e. we create an array traj_idx in which traj_idx[i] is the starting index of the data for the (i+1)th trajectory (since Python uses zero-based indexing.).
traj_idx = np.insert(np.cumsum(ds.rowsize.values), 0, 0)
Now, we can retrieve a given variable of a specified trajectory but note that we need to indicate both the start and the end indices of the data for that trajectory. For example, get the SST time series for the 11th trajectory:
i = 10
# .compute() is necessary here to explicitely load the data from the file and execute the operation
ds.sst[slice(traj_idx[i], traj_idx[i+1])].compute()
<xarray.DataArray 'sst' (obs: 4151)>
array([ nan, 290.941, 290.967, ..., 281.062, 281.037, nan],
dtype=float32)
Coordinates:
lon (obs) float32 -55.01 -55.01 -55.01 -55.0 ... -25.87 -25.85 -25.84
lat (obs) float32 -39.96 -39.95 -39.95 -39.94 ... -45.79 -45.78 -45.78
time (obs) datetime64[ns] 2002-12-18T19:00:00 ... 2003-06-13T07:00:00
ids (obs) int64 27131 27131 27131 27131 ... 27131 27131 27131 27131
Dimensions without coordinates: obs
Attributes:
long_name: Fitted sea water temperature
units: Kelvin
comments: Estimated near-surface sea water temperature from drifting bu...The same sst variable can be retrieved by using the drifter’s ID:
id_ex = ds.ID[i].values # id of the i+1 th trajectory as an example
id_ex
array(27131)
j = int(np.where(ds.ID == id_ex)[0]) # retrieve back the index
ds.sst[slice(traj_idx[j], traj_idx[j+1])].compute()
<xarray.DataArray 'sst' (obs: 4151)>
array([ nan, 290.941, 290.967, ..., 281.062, 281.037, nan],
dtype=float32)
Coordinates:
lon (obs) float32 -55.01 -55.01 -55.01 -55.0 ... -25.87 -25.85 -25.84
lat (obs) float32 -39.96 -39.95 -39.95 -39.94 ... -45.79 -45.78 -45.78
time (obs) datetime64[ns] 2002-12-18T19:00:00 ... 2003-06-13T07:00:00
ids (obs) int64 27131 27131 27131 27131 ... 27131 27131 27131 27131
Dimensions without coordinates: obs
Attributes:
long_name: Fitted sea water temperature
units: Kelvin
comments: Estimated near-surface sea water temperature from drifting bu...Once we know the indices of the trajectory of interest, we can calculate statistics like mean() or other reducing operations on one, or a list of variables:
s_var = ['ve', 'vn', 'gap', 'err_lat', 'err_lon', 'sst', 'sst1', 'sst2', 'err_sst', 'err_sst1', 'err_sst2']
ds.isel({'obs': slice(traj_idx[i], traj_idx[i+1]), 'traj': i})[s_var].mean().compute()
<xarray.Dataset>
Dimensions: ()
Coordinates:
ID int64 27131
Data variables:
ve float32 0.1437
vn float32 -0.04261
gap float32 1.479e+04
err_lat float32 0.007155
err_lon float32 0.01117
sst float32 285.7
sst1 float32 285.7
sst2 float32 -0.003311
err_sst float32 0.07059
err_sst1 float32 0.02489
err_sst2 float32 0.06399Operations per trajectory#
As previously noted, to perform the same operation per trajectory, it is more efficient to align the chunks with the trajectories. This is done by settings the size of the chunks equal to the dimensions of the trajectories. However, here we need to know the lengths of all the trajectories in the order they are arranged in the ragged array before actually loading the dataset with xarray. This is clearly not practical as xarray does not provide an obvious mechanism to align the data chunks with specified indices.
rowsize = np.zeros(len(files), dtype='int')
for i, file in enumerate(files):
with xr.open_dataset(file, decode_times=False) as ds_t:
rowsize[i] = ds_t.sizes['obs']
ds.close() # close previously loaded xr.Dataset
# align chunks of the dask array with the length of each individual trajectory
chunk_settings = {'obs': tuple(rowsize.tolist())}
ds = xr.open_dataset('data/gdp_subset.nc', chunks=chunk_settings)
Variables such as the longitude, are now split into subset_nb_drifters chunks which have different sizes and where the maximum size is indicated by the Shape characteristic of the xarray.DataArray objects. As an example for lon:
ds.lon
<xarray.DataArray 'lon' (obs: 4786301)>
dask.array<open_dataset-7f2a95543265e3b414ce19877f3d517clon, shape=(4786301,), dtype=float32, chunksize=(50655,), chunktype=numpy.ndarray>
Coordinates:
lon (obs) float32 dask.array<chunksize=(1095,), meta=np.ndarray>
lat (obs) float32 dask.array<chunksize=(1095,), meta=np.ndarray>
time (obs) datetime64[ns] dask.array<chunksize=(1095,), meta=np.ndarray>
ids (obs) int64 dask.array<chunksize=(1095,), meta=np.ndarray>
Dimensions without coordinates: obs
Attributes:
long_name: Longitude
units: degrees_eastNaturally, now loading a trajectory always require loading only one chunk.
i = 10
ds.lon.isel({'obs': slice(traj_idx[i], traj_idx[i+1])})
<xarray.DataArray 'lon' (obs: 4151)>
dask.array<getitem, shape=(4151,), dtype=float32, chunksize=(4151,), chunktype=numpy.ndarray>
Coordinates:
lon (obs) float32 dask.array<chunksize=(4151,), meta=np.ndarray>
lat (obs) float32 dask.array<chunksize=(4151,), meta=np.ndarray>
time (obs) datetime64[ns] dask.array<chunksize=(4151,), meta=np.ndarray>
ids (obs) int64 dask.array<chunksize=(4151,), meta=np.ndarray>
Dimensions without coordinates: obs
Attributes:
long_name: Longitude
units: degrees_eastSimple operation by chunks (or blocks)#
To perform arbitrary operation per trajectory, we can now use map_blocks() that applies a function per chunk (block is also used for chunks). In the example below we simply define the function func that calculates the mean of each block of an array x. Note that this requires loading nb_subset_drifters chunks and applying the operation.
def func(x):
return np.array([np.nanmean(x)])
stats_traj = ds.sst.data.map_blocks(func).compute() # apply to sst variable
/var/folders/jh/92r7zqw159xbv25hk5fbrzgw0000gp/T/ipykernel_2540/1074368307.py:2: RuntimeWarning: Mean of empty slice
return np.array([np.nanmean(x)])
stats_traj[:10]
array([275.79956, 280.2327 , 277.82953, 282.86588, 293.74094, 293.33972,
287.9495 , 271.2664 , 293.4779 , 300.82187], dtype=float32)
More complex operation where dimensions can change between input and output#
Alternatively, one can use the xarray function apply_ufunc to map a function per chunks. In this case, the function to be applied can be a bit more complex, but this method gives more flexibility to handle the input and output formats and dimensions of the function.
In a first example, the eastward velocity is passed as an argument by chunk to the function which will calculate the anomaly per trajectory. The length of the output will be of the same length as the input. Please note here that:
The input and output sizes are not modified.
An argument must be passed to the function. It shouldn’t be
ds.ve(the xarray DataArray), but theds.ve.data(the underlying dask core data array) must be used.
def per_chunk_anomaly(array):
# MUST return an array (https://github.com/dask/dask/issues/8822)
return array.map_blocks(lambda x: x-np.nanmean(x), chunks=array.chunks)
# here the apply_ufunc functions takes the function just defined above(per_chunk_anomaly)
# and the data to which apply that function
ret = xr.apply_ufunc(
per_chunk_anomaly,
ds.ve.data, # .data to retrieve the underlying chunked dask array
input_core_dims=[['obs']],
output_core_dims=[['obs']],
dask='allowed'
).compute()
And display some results:
display(ret) # only a few values will be shown by default
display(len(ret)) # length of the output should be same as length of input
array([0.02569735, 0.02569735, 0.02599735, ..., 0.13741829, 0.13441828,
0.12031829], dtype=float32)
4786301
In a second example, we calculates mean() values per trajectory. This means that for our datasets consisting of nb_subset_drifters chunks, ttput will have nb_subset_drifters values, that is one value per chunk. In other words, the length of the output will be different from the length of the input.
def per_chunk_mean(array):
nchunks = len(array.chunks[0])
output_chunks = ([1] * nchunks,) # 1 value per chunk
return array.map_blocks(lambda x: np.nanmean(x, keepdims=True), chunks=output_chunks)
ret = xr.apply_ufunc(
per_chunk_mean,
ds.ve.data,
input_core_dims=[['obs']], # input dimension to the per_block function
output_core_dims=[['obs']], # output still has one dimension
exclude_dims=set(['obs']), # size of x changes so it has to be in the exclude_dims param
dask='allowed',
).compute()
And display some results:
display(ret[:10]) # show only first 10 values
display(len(ret)) # length of output should be the number of trajectories
array([-0.03099735, 0.14964446, 0.152546 , 0.15595299, 0.01793322,
-0.01830383, 0.03096705, -0.03741267, -0.00138449, 0.03001278],
dtype=float32)
500
In a last example, we estimate ocean velocity rotary spectra of the complex-valued time series u+vi via the periodogram method (and using the NumPy fft function). Here, the combination of the two methods apply_ufunc and map_blocks is a powerful tool which can serve as templates for a range of advanced processing applied per trajectory.
First, we create the new complex-valued variable cv which inherits the chunks from ve and vn:
ds = ds.assign(cv = ds['ve'] + 1j*ds['vn'])
ds.cv
<xarray.DataArray 'cv' (obs: 4786301)>
dask.array<add, shape=(4786301,), dtype=complex64, chunksize=(50655,), chunktype=numpy.ndarray>
Coordinates:
lon (obs) float32 dask.array<chunksize=(1095,), meta=np.ndarray>
lat (obs) float32 dask.array<chunksize=(1095,), meta=np.ndarray>
time (obs) datetime64[ns] dask.array<chunksize=(1095,), meta=np.ndarray>
ids (obs) int64 dask.array<chunksize=(1095,), meta=np.ndarray>
Dimensions without coordinates: obsdef per_chunk_periodogram(array):
dt = 1/24 # [-]
return array.map_blocks(lambda x: dt*np.abs(np.fft.fft(x))**2, chunks=array.chunks)
Here we benchmark this last, more complex, operation:
t0 = time.time()
ret = xr.apply_ufunc(
per_chunk_periodogram,
ds['cv'].data,
input_core_dims=[["obs"]], # input dimension to the per_block function
output_core_dims=[["obs"]], # output still has one dimension
dask='allowed'
).compute()
benchmark_times[0,2] = time.time() - t0
The return variable is the same length as ds['cv'], and is a ragged array of estimated rotary spectra by the periodogram method.
(Please note: the periodogram is not a good spectral estimator at all and alternative methods such as multitaper methods should be used to estimate spectra. Please also note that we assumed that the individual trajectories were gap-free at regular hourly intervals which is required to apply a FFT. However, some velocity time series have gaps, in which case the FFT calculation is not valid)
Display one example spectrum:
i = 0 # drifter index
# calculate inertial frequency in cycle per day [cpd] from the mean latitude of trajectory
omega = 7.2921159e-5 # Earth's rotation rate [rad/s]
seconds_per_day = 60*60*24 # [s]
fi = -2*omega*(seconds_per_day/(2*np.pi))*np.sin(np.radians(ds.lat[traj_idx[i]:traj_idx[i+1]]).mean().compute())
# define frequency scale/abscissa
dt = 1/24 # [-]
f = np.fft.fftfreq(int(ds.rowsize[i].compute()), dt)
ss = np.log10(ret[traj_idx[i]:traj_idx[i+1]]) # spectrum of the ith trajectory
fig = plt.figure(dpi=150)
ax = fig.add_subplot(1,1,1)
h1, = ax.plot(np.fft.fftshift(f), np.fft.fftshift(ss), scaley=True, label='Spectrum', linewidth=0.5)
h2 = ax.axvline(x=fi,color='k',linestyle=':', label='Inertial frequency', linewidth=0.5)
ax.legend(frameon=False)
ax.set_ylabel('PSD (m$^2$ s$^{-2}$ cpd$^{-1}$)')
ax.set_xlabel('Frequency [cpd]');
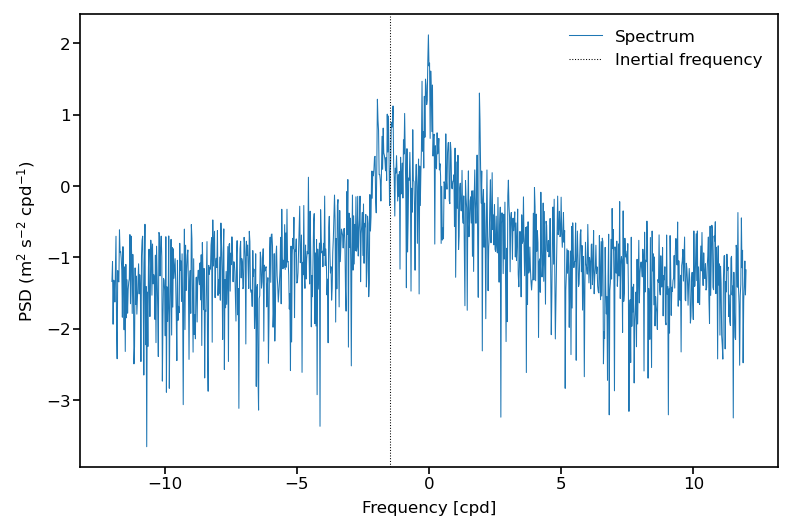
At this stage it is necessary to clear the memory (RAM) of the Python process because in the next step, the data are fully loaded in memory by using Pandas.
del ds, ds_subset, ret, traj_idx, stats_traj, rowsize, x_c, y_c
Pandas#
In this section, we test the use of the Pandas package for our Lagrangian data. Since the previously-created NetCDF file containing the ragged array has two dimensions ['traj'] and ['obs'], it cannot be read directly into the table structure of a pandas.DataFrame(). As a workaround, we load the data with xarray, drop the ['traj'] dimension, and convert the resulting data to a pandas.DataFrame(). The disadvantage is that it requires to form a second dataset with the dimension['traj'] to have access to all variables (not presented here).
df = xr.open_dataset('data/gdp_subset.nc').drop_dims('traj').to_pandas() # only contains the variables with dimension ['obs']
In this case, the data is organized in a table format where each variable is associated with a column, and each row contains one observation. A DataFrame is easy to quickly look at, with the .head(), .tail() functions as an example:
df.head()
| ve | vn | gap | err_lat | err_lon | err_ve | err_vn | drogue_status | sst | sst1 | ... | err_sst | err_sst1 | err_sst2 | flg_sst | flg_sst1 | flg_sst2 | lon | lat | time | ids | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| obs | |||||||||||||||||||||
| 0 | -0.0053 | 0.0004 | 15638.0 | 0.00391 | 0.00797 | 0.0650 | 0.0279 | False | 273.471985 | 274.126007 | ... | 0.074 | 0.224 | 0.236 | 5 | 5 | 4 | -52.393002 | 47.307461 | 2001-05-01 08:00:00 | 2592 |
| 1 | -0.0053 | 0.0007 | 15638.0 | 0.00345 | 0.00676 | 0.0603 | 0.0284 | False | 273.100006 | 273.760010 | ... | 0.077 | 0.158 | 0.185 | 5 | 5 | 4 | -52.393070 | 47.307590 | 2001-05-01 09:00:00 | 2592 |
| 2 | -0.0050 | 0.0015 | 15638.0 | 0.00313 | 0.00586 | 0.0548 | 0.0290 | False | 272.869995 | 273.513000 | ... | 0.086 | 0.113 | 0.142 | 5 | 5 | 4 | -52.392960 | 47.307861 | 2001-05-01 10:00:00 | 2592 |
| 3 | -0.0037 | 0.0035 | 15638.0 | 0.00288 | 0.00503 | 0.0511 | 0.0297 | False | 272.752991 | 273.334015 | ... | 0.096 | 0.089 | 0.123 | 5 | 5 | 4 | -52.392208 | 47.308449 | 2001-05-01 11:00:00 | 2592 |
| 4 | -0.0044 | 0.0031 | 13651.0 | 0.00292 | 0.00475 | 0.0509 | 0.0313 | False | 272.743988 | 273.213013 | ... | 0.102 | 0.078 | 0.118 | 5 | 5 | 4 | -52.391151 | 47.309021 | 2001-05-01 12:00:00 | 2592 |
5 rows × 21 columns
print(f'This subset contains {len(np.unique(df.ids))} drifters and {len(df):,} observations.')
This subset contains 500 drifters and 4,786,301 observations.
Pandas test 1: Geographical binning of any variable#
Here we reconduct the first bench test: calculate the mean zonal velocity from all drifters of the dataset.
t0 = time.time()
lon = np.linspace(-180, 180, 360*2)
lat = np.linspace(-90, 90, 180*2)
ret = stats.binned_statistic_2d(df.lon,
df.lat,
df.ve,
statistic=np.nanmean, # can pass any function()
bins=[lon, lat])
benchmark_times[1,0] = time.time() - t0
x_c = np.convolve(lon, [0.5, 0.5], mode='valid')
y_c = np.convolve(lat, [0.5, 0.5], mode='valid')
# get 1st and 99th percentiles of values to plot to get a useful range for the colorscale
v1,v2 = np.nanpercentile(ret.statistic.T,[1,99])
fig = plt.figure(dpi=150)
ax = fig.add_subplot(1,1,1,projection=ccrs.Robinson(central_longitude=-180))
cmap = cmocean.tools.crop(cmocean.cm.balance, vmin=v1, vmax=v2, pivot=0)
pcm = ax.pcolormesh(x_c, y_c,
ret.statistic.T,
cmap=cmap,
transform=ccrs.PlateCarree(),
vmin=v1, vmax=v2)
# gridlines and labels
gl = ax.gridlines(color='k', linewidth=0.1, linestyle='-',
xlocs=np.arange(-180, 181, 60), ylocs=np.arange(-90, 91, 30),
draw_labels=True)
gl.top_labels = False
gl.right_labels = False
# add land and coastline
ax.add_feature(cfeature.LAND, facecolor='grey', zorder=1)
ax.add_feature(cfeature.COASTLINE, linewidth=0.25, zorder=1)
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", size="3%", pad=0.05, axes_class=plt.Axes)
cb = fig.colorbar(pcm, cax=cax);
cb.ax.set_ylabel('Zonal velocity [m/s]');
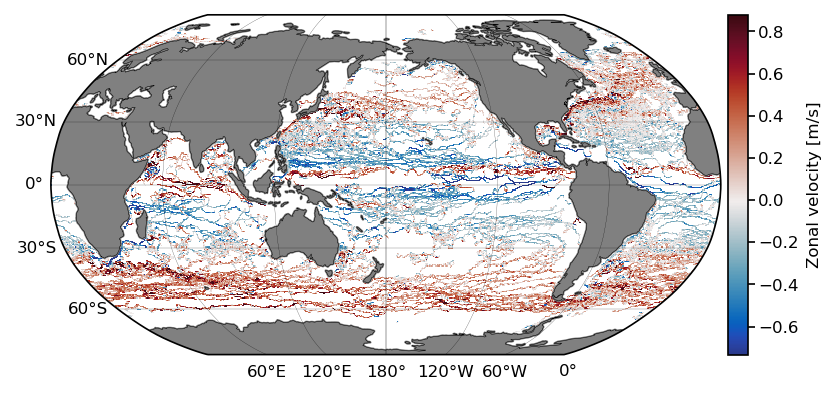
Pandas test 2: Extract a given region#
The extraction of a region is slightly simpler with Pandas than with xarray, because the same mask can be applied to all variables of interest that share the obs dimension.
def retrieve_region_pd(df, lon: list = None, lat: list = None, time: list = None):
'''Subset the dataset for a region in space and time
Args:
df: Pandas DataFrame
lon: longitude slice of the subregion
lat: latitude slice of the subregion
time: tiem slice of the subregion
Returns:
ds_subset: Dataset of the subregion
'''
mask = functools.reduce(np.logical_and,
(
df.lon >= lon[0],
df.lon <= lon[1],
df.lat >= lat[0],
df.lat <= lat[1],
df.time >= day0,
df.time <= day1,
)
)
return df.loc[mask]
t0 = time.time()
lon = [-98, -78]
lat = [18, 31]
day0 = datetime(2000,1,1)
day1 = datetime(2020,12,31)
days = [day0, day1]
df_subset = retrieve_region_pd(df, lon, lat, days)
benchmark_times[1,1] = time.time() - t0
df_subset.head()
| ve | vn | gap | err_lat | err_lon | err_ve | err_vn | drogue_status | sst | sst1 | ... | err_sst | err_sst1 | err_sst2 | flg_sst | flg_sst1 | flg_sst2 | lon | lat | time | ids | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| obs | |||||||||||||||||||||
| 902352 | -0.3192 | -0.0632 | 10886.0 | 0.00611 | 0.01212 | 0.1262 | 0.0633 | False | 301.339996 | 301.382996 | ... | 0.022 | 0.007 | 0.019 | 5 | 5 | 5 | -78.007187 | 30.198179 | 2006-09-15 14:00:00 | 54388 |
| 902353 | -0.4248 | -0.0653 | 10886.0 | 0.00193 | 0.01652 | 0.1244 | 0.0656 | False | 301.377014 | 301.386993 | ... | 0.021 | 0.007 | 0.018 | 5 | 5 | 2 | -78.036041 | 30.196760 | 2006-09-15 15:00:00 | 54388 |
| 902354 | -0.1182 | -0.0115 | 4925.0 | 0.00219 | 0.01249 | 0.1813 | 0.0548 | False | 301.417999 | 301.390015 | ... | 0.018 | 0.006 | 0.016 | 5 | 5 | 5 | -78.044693 | 30.193790 | 2006-09-15 16:00:00 | 54388 |
| 902355 | -0.0863 | -0.0425 | 1210.0 | 0.00063 | 0.00932 | 0.1501 | 0.0463 | False | 301.457001 | 301.391998 | ... | 0.015 | 0.006 | 0.014 | 5 | 5 | 5 | -78.048553 | 30.192690 | 2006-09-15 17:00:00 | 54388 |
| 902356 | 0.0080 | -0.0562 | 4925.0 | 0.00415 | 0.00690 | 0.1936 | 0.0922 | False | 301.487000 | 301.394012 | ... | 0.013 | 0.005 | 0.012 | 5 | 5 | 5 | -78.045929 | 30.191130 | 2006-09-15 18:00:00 | 54388 |
5 rows × 21 columns
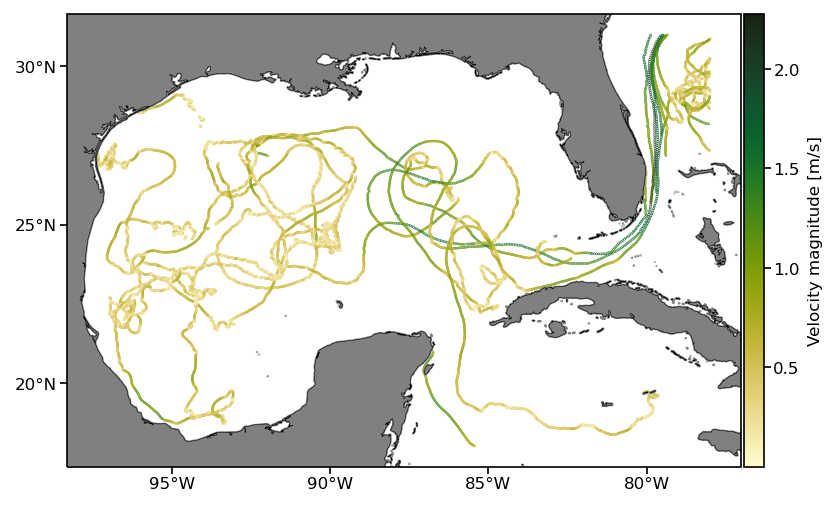
Here we plot the result as before but displaying the speed for each data point:
fig = plt.figure(dpi=150)
ax = fig.add_subplot(1,1,1,projection=ccrs.PlateCarree())
pcm = ax.scatter(df_subset.lon, df_subset.lat,
s=0.05, c=np.sqrt(df_subset.ve**2+df_subset.vn**2), transform=ccrs.PlateCarree(),
cmap=cmocean.cm.speed)
ax.add_feature(cfeature.LAND, facecolor='grey', zorder=1)
ax.add_feature(cfeature.COASTLINE, linewidth=0.25, zorder=1)
ax.set_xticks(np.arange(-95, -79, 5), crs=ccrs.PlateCarree())
ax.set_yticks([20, 25, 30], crs=ccrs.PlateCarree())
ax.xaxis.set_major_formatter(LongitudeFormatter())
ax.yaxis.set_major_formatter(LatitudeFormatter())
divider = make_axes_locatable(ax)
cax = divider.append_axes('right', size='3%', pad=0.02, axes_class=plt.Axes)
cb = fig.colorbar(pcm, cax=cax)
cb.set_label('Velocity magnitude [m/s]')
Pandas test 3: Single statistic per trajectory#
Single operation per trajectory can be easily implemented in a Pandas DataFrame by locating the drifter ID by index (iloc) or by value (loc).
i = 10 # drifter index
df.iloc[np.where(df['ids'] == np.unique(df['ids'])[i])].mean(numeric_only=False)
ve 0.143713
vn -0.042606
gap 14790.94043
err_lat 0.007155
err_lon 0.01117
err_ve 0.095038
err_vn 0.064061
drogue_status 0.998073
sst 285.678101
sst1 285.681427
sst2 -0.003311
err_sst 0.070594
err_sst1 0.024895
err_sst2 0.063991
flg_sst 4.997591
flg_sst1 4.997591
flg_sst2 3.857866
lon -45.807869
lat -45.521275
time 2003-03-17 05:04:36.656227328
ids 27131.0
dtype: object
Assuming we know a drifter ID id_ex (here we simply use the value associated to the ith index above), we can retrieve the associated data by value as follow.
id_ex = np.unique(df['ids'])[i]
id_ex
27131
df.loc[df['ids'] == id_ex].mean(numeric_only=False)
ve 0.143713
vn -0.042606
gap 14790.94043
err_lat 0.007155
err_lon 0.01117
err_ve 0.095038
err_vn 0.064061
drogue_status 0.998073
sst 285.678101
sst1 285.681427
sst2 -0.003311
err_sst 0.070594
err_sst1 0.024895
err_sst2 0.063991
flg_sst 4.997591
flg_sst1 4.997591
flg_sst2 3.857866
lon -45.807869
lat -45.521275
time 2003-03-17 05:04:36.656227328
ids 27131.0
dtype: object
For more complicated operations per trajectory, the pandas groupby() function allows us to split the DataFrame. Applied to the ids['obs'] (which repeat the ID of a drifter for each of its observations), we can obtain a grouped object containing one trajectory per group. Following this operation, one can apply a predefined set of operations, such as mean(), or any user defined functions using .apply(). As before, we provide an example where we can calculate the mean of SST per trajectory, and another example of estimating velocity spectra by the periodogram method.
ret = df.groupby("ids").sst.mean()
ret
ids
2592 275.799561
6428 280.232697
13566 277.829529
17927 282.865875
18706 293.740936
...
68243870 273.882111
68244460 273.744598
68244730 273.515564
68246720 296.040009
68248530 286.112793
Name: sst, Length: 500, dtype: float32
Again, to estimate the velocity spectra we have to create the new complex-valued variable cv from ve and vn:
df['cv'] = df['ve'] + 1j*df['vn']
And calculate the periodogram per trajectory using this new variable:
def periodogram_per_group(array):
dt = 1/24
return dt*np.abs(np.fft.fft(array))**2
t0 = time.time()
ret = df.groupby("ids").cv.apply(periodogram_per_group)
benchmark_times[1,2] = time.time() - t0
This time, the results are stored into a dictionary where the keys are the drifters’ ID:
ret
ids
2592 [48.01623216121766, 52.74661085729767, 4.63967...
6428 [341830.5755422627, 53330.50826245666, 857.134...
13566 [46152.48004791965, 6588.974982702436, 1458.62...
17927 [325605.62376754, 63676.24022864982, 11794.261...
18706 [1076.965552542786, 773.9372866187755, 311.397...
...
68243870 [127.7695912061453, 215.20477509848652, 315.08...
68244460 [301.4939495649164, 64.02978051130118, 424.069...
68244730 [383.1069530335443, 67.31997165755263, 7.50842...
68246720 [308.7263083474708, 1125.3042873617292, 4771.3...
68248530 [49.898529559024354, 2.2134116883119983, 15.66...
Name: cv, Length: 500, dtype: object
As an example, we display the first 10 values of the sprctrum for drifter id_ex:
ret[id_ex][:10]
array([16131.45017614, 16040.80300053, 3017.6902432 , 6700.41755415,
311.93297982, 1555.5981903 , 757.26867001, 7200.28268359,
891.88959794, 1036.35997949])
del df, df_subset, ret, x_c, y_c # again free the memory before the next section
Awkward Array#
Awkward Array is a library for nested, variable-sized data, including arbitrary-length lists, records, mixed types, and missing data, using NumPy-like idioms. Such arrays are dynamically typed (defined at runtime as a function of the variable to store), but operations on them are compiled into machine code and therefore executes faster than regular interpreted Python function. This allow to generalizes NumPy array manipulation routines, even with variable-sized data.
A schematic view of the data structure is presented in the figure below. The higher level ak.Array and ak.Record hides the nested structure of the data structure, but can always be accessed with ak.Array.layout (or ds_ak.layout in our case). This structure also stores parameters or metadata such as units, long name, etc. associated with the data. The library stores data into standard NumPyArray alongside a ListOffsetArray for fast and efficient access to variables, even with nonuniform data length.

The main advantages of Awkward Array are:
the indexing is already taken care of by the library, e.g.
sst[400]returns the sea surface temperature along the 401st trajectory;ability to efficiently perform operations per trajectory;
easy integration with Numba (demonstrated below in section 7.4).
An Awkward array can easily be created by reading our NetCDF file:
ds_ak = create_ak(xr.open_dataset('data/gdp_subset.nc', decode_times=False))
Awkward Arrays supports N-dimensional datasets by storing nested arrays. The main dataset contains variables (type = subset_nb_drifters * {various type}) containing the metadata.
ds_ak
<Array [{ID: 2592, rowsize: 1095, ... ] type='500 * struct[["ID", "rowsize", "lo...'>
Awkward Array doesn’t (yet!) have an html representation of its structure, but it is possible to list the fields.
ds_ak.fields # list of fields/variables
['ID',
'rowsize',
'location_type',
'WMO',
'expno',
'deploy_date',
'deploy_lat',
'deploy_lon',
'end_date',
'end_lat',
'end_lon',
'drogue_lost_date',
'type_death',
'type_buoy',
'DeploymentShip',
'DeploymentStatus',
'BuoyTypeManufacturer',
'BuoyTypeSensorArray',
'CurrentProgram',
'PurchaserFunding',
'SensorUpgrade',
'Transmissions',
'DeployingCountry',
'DeploymentComments',
'ManufactureYear',
'ManufactureMonth',
'ManufactureSensorType',
'ManufactureVoltage',
'FloatDiameter',
'SubsfcFloatPresence',
'DrogueType',
'DrogueLength',
'DrogueBallast',
'DragAreaAboveDrogue',
'DragAreaOfDrogue',
'DragAreaRatio',
'DrogueCenterDepth',
'DrogueDetectSensor',
'obs']
If we peak inside one of the field (or variable), we can see the associated type. For example, the drifters’ ID have a type=subset_nb_drifters * int64.
ds_ak.ID # one variable
<Array [2592, 6428, ... 68246720, 68248530] type='500 * int64[parameters={"attrs...'>
If we display the last field ‘obs’, we can see that it is a nested Awkward Array and contains all variables with type=(subset_nb_drifters * var * dtype) containing data along trajectories.
ds_ak.obs
<Array [{lon: [-52.4, -52.4, -52.4, ... 5, 5]}] type='500 * {"lon": [var * float...'>
The library includes the function ak.flatten(), similar to np.flatten() to concatenate all observations for one variable into a unidimensional array. As an example, with ‘sst’:
print(f'Total length of the data inside \'obs\' field \'sst\' are {len(ak.flatten(ds_ak.obs.sst)):,}.')
ak.flatten(ds_ak.obs.sst)
Total length of the data inside 'obs' field 'sst' are 4,786,301.
<Array [273, 273, 273, 273, ... 287, 288, 288] type='4786301 * float32'>
A nested structure is utilized to efficiently store N-dimensional dataset, in our case we first have the fields with dimension ['traj'], followed by the fields with dimension ['obs'] stored inside the array ds_ak.obs.
ds_ak.obs.fields
['lon',
'lat',
'time',
'ids',
've',
'vn',
'gap',
'err_lat',
'err_lon',
'err_ve',
'err_vn',
'drogue_status',
'sst',
'sst1',
'sst2',
'err_sst',
'err_sst1',
'err_sst2',
'flg_sst',
'flg_sst1',
'flg_sst2']
For example, the type of the sst field, stored at ds_ak.obs.sst, is 17324 * var * float32. This means that there are 17324 trajectories of variable size of float32`.
ds_ak.obs.sst
<Array [[273, 273, 273, ... 287, 288, 288]] type='500 * [var * float32, paramete...'>
The data along the trajectory of a specific drifter can easily be retrieved using an index.
i = 0
ds_ak.obs.sst[i]
<Array [273, 273, 273, 273, ... 280, 280, nan] type='1095 * float32'>
The same data can be extracted from the drifter ID:
id_ex = ds_ak.ID[i] # id of the ith trajectory as an example
display(id_ex)
i = np.where(ds_ak.ID == id_ex)[0][0]
ds_ak.obs.sst[i]
2592
<Array [273, 273, 273, 273, ... 280, 280, nan] type='1095 * float32'>
And the data along a specific trajectory can be accessed using a second index. As an example, the first sst value of the ith trajectory:
ds_ak.obs.sst[i][0] # [K]
273.47198486328125
Metadata or attributes are store for the global Array ds_ak.layout.parameters, as well as for each individual variable (ds_ak.obs.sst.layout.parameters), similarly to global attributes and variable attributes in a NetCDF file. Those attributes are stored as a parameter in the ak.RecordArray (obtained with the .layout member), which summarizes the data fields, the types, and the shapes of the data itself.
ds_ak.layout.parameters
{'attrs': {'title': 'Global Drifter Program hourly drifting buoy collection',
'history': 'Version 2.00. Metadata from dirall.dat and deplog.dat',
'Conventions': 'CF-1.6',
'date_created': '2022-05-20T14:48:40.763710',
'publisher_name': 'GDP Drifter DAC',
'publisher_email': 'aoml.dftr@noaa.gov',
'publisher_url': 'https://www.aoml.noaa.gov/phod/gdp',
'licence': 'MIT License',
'processing_level': 'Level 2 QC by GDP drifter DAC',
'metadata_link': 'https://www.aoml.noaa.gov/phod/dac/dirall.html',
'contributor_name': 'NOAA Global Drifter Program',
'contributor_role': 'Data Acquisition Center',
'institution': 'NOAA Atlantic Oceanographic and Meteorological Laboratory',
'acknowledgement': 'Elipot et al. (2022) to be submitted. Elipot et al. (2016). Global Drifter Program quality-controlled hourly interpolated data from ocean surface drifting buoys, version 2.00. NOAA National Centers for Environmental Information. https://agupubs.onlinelibrary.wiley.com/doi/full/10.1002/2016JC011716TBA. Accessed [date].',
'summary': 'Global Drifter Program hourly data'}}
ds_ak.obs.sst.layout.parameters
{'attrs': {'long_name': 'Fitted sea water temperature',
'units': 'Kelvin',
'comments': 'Estimated near-surface sea water temperature from drifting buoy measurements. It is the sum of the fitted near-surface non-diurnal sea water temperature and fitted diurnal sea water temperature anomaly. Discrepancies may occur because of rounding.'}}
Awkward Array test 1: Geographical binning of any variable#
Again, we reconduct the first bench test: calculate the mean zonal velocity from all drifters of the dataset. For operations apply on the full dataset, we use the ak.flatten() function to retrieve the underlying unidimensional NumPy array containing all the observations for all drifters.
t0 = time.time()
lon = np.linspace(-180, 180, 360 * 2)
lat = np.linspace(-90, 90, 180 * 2)
ret = stats.binned_statistic_2d(ak.flatten(ds_ak.obs.lon, axis=1),
ak.flatten(ds_ak.obs.lat, axis=1),
ak.flatten(ds_ak.obs.ve, axis=1),
statistic=np.nanmean,
bins=[lon, lat])
benchmark_times[2,0] = time.time() - t0
x_c = np.convolve(lon, [0.5, 0.5], mode='valid')
y_c = np.convolve(lat, [0.5, 0.5], mode='valid')
# get 1st and 99th percentiles of values to plot to get a useful range for the colorscale
v1,v2 = np.nanpercentile(ret.statistic.T,[1,99])
fig = plt.figure(dpi=150)
ax = fig.add_subplot(1,1,1,projection=ccrs.Robinson(central_longitude=-180))
cmap = cmocean.tools.crop(cmocean.cm.balance, vmin=v1, vmax=v2, pivot=0)
pcm = ax.pcolormesh(x_c, y_c,
ret.statistic.T,
cmap=cmap,
transform=ccrs.PlateCarree(),
vmin=v1, vmax=v2)
# gridlines and labels
gl = ax.gridlines(color='k', linewidth=0.1, linestyle='-',
xlocs=np.arange(-180, 181, 60), ylocs=np.arange(-90, 91, 30),
draw_labels=True)
gl.top_labels = False
gl.right_labels = False
# add land and coastline
ax.add_feature(cfeature.LAND, facecolor='grey', zorder=1)
ax.add_feature(cfeature.COASTLINE, linewidth=0.25, zorder=1)
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", size="3%", pad=0.05, axes_class=plt.Axes)
cb = fig.colorbar(pcm, cax=cax);
cb.ax.set_ylabel('Zonal velocity [m/s]');
Awkward Array test 2: Extract a given region#
With Awkward Array, the extraction of a subregion is slightly more complex than with Pandas, since here the dataset contains both variables with dimension ['traj'] and variables with dimension ['obs']. Note that comparisons can be performed on the bidimensional non-regular ak.Array (e.g. ds.obs.lon, ds.obs.lat, ds.obs.time) without looping.
def retrieve_region_ak(ds: ak.Array, lon: list = None, lat: list = None, days: list = None) -> ak.Array:
'''Subset the dataset for a region in space and time
Args:
ds: Awkward Array
lon: longitude slice of the subregion
lat: latitude slice of the subregion
days: days slice of the subregion
Returns:
ds_subset: Dataset of the subregion
'''
mask = functools.reduce(np.logical_and,
(
ds.obs.lon >= lon[0],
ds.obs.lon <= lon[1],
ds.obs.lat >= lat[0],
ds.obs.lat <= lat[1],
ds.obs.time >= days[0],
ds.obs.time <= days[1],
)
)
ds_s = ak.copy(ds)
mask_id = np.in1d(ds_s.ID, np.unique(ak.flatten(ds_s.obs.ids[mask])))
ds_s = ds_s[mask_id] # mask for variables with dimension ['traj']
ds_s.obs = ds_s.obs[mask[mask_id]] # mask for variables with dimension ['obs']
return ds_s
t0 = time.time()
lon = [-98, -78]
lat = [18, 31]
day0 = (datetime(2000,1,1) - datetime(1970,1,1)).total_seconds()
day1 = (datetime(2020,12,31) - datetime(1970,1,1)).total_seconds()
days = [day0, day1]
ds_subset = retrieve_region_ak(ds_ak, lon, lat, days)
benchmark_times[2,1] = time.time() - t0
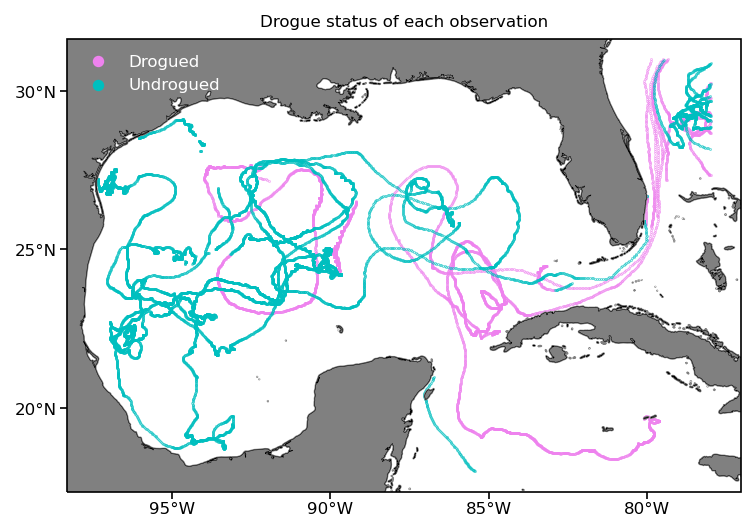
Here we plot the result as before but displaying the drogue status of drifters along their trajectory. Note: the drogue is a sea anchor centered at 15 m depth that can become detached, after which the drifter is more strongly affected by surface winds and waves.
fig = plt.figure(dpi=150)
ax = fig.add_subplot(1,1,1,projection=ccrs.PlateCarree())
cond = ds_subset.obs.drogue_status # True: drogued, False: undrogued
pcm = ax.scatter(ak.flatten(ds_subset.obs.lon[cond]), ak.flatten(ds_subset.obs.lat[cond]),
s=0.05, c='violet', transform=ccrs.PlateCarree(), label='Drogued')
pcm = ax.scatter(ak.flatten(ds_subset.obs.lon[~cond]), ak.flatten(ds_subset.obs.lat[~cond]),
s=0.05, c='c', transform=ccrs.PlateCarree(), label='Undrogued')
ax.add_feature(cfeature.LAND, facecolor='grey', zorder=1)
ax.add_feature(cfeature.COASTLINE, linewidth=0.25, zorder=1)
ax.set_xticks(np.arange(-95, -79, 5), crs=ccrs.PlateCarree())
ax.set_yticks([20, 25, 30], crs=ccrs.PlateCarree())
ax.xaxis.set_major_formatter(LongitudeFormatter())
ax.yaxis.set_major_formatter(LatitudeFormatter())
ax.set_title('Drogue status of each observation', size=8);
ax.legend(markerscale=20, labelcolor='w', frameon=False);
Awkward Array test 3: Single statistic per trajectory#
Without the need for indices, we can perform reduction operation per trajectory directly with Awkward array, which simplify considerably the notations, and should ease the development of new functionality.
Note: At this point, the dataset ds_ak is loaded in memory, which explains the speed of the operations. For larger dataset that cannot be held completely in memory, an option could be to interface with Dask, currently under development.
avg_sst = np.zeros(len(ds_ak.obs))
for i, s in enumerate(ds_ak.obs.sst):
avg_sst[i] = np.nanmean(s)
avg_sst[:10]
/Users/pmiron/miniforge3/envs/research/lib/python3.9/site-packages/awkward/_connect/_numpy.py:36: RuntimeWarning: Mean of empty slice
out = func(*args, **kwargs)
array([275.79956055, 280.23269653, 277.82952881, 282.86587524,
293.74093628, 293.33972168, 287.94949341, 271.26638794,
293.47790527, 300.8218689 ])
For basic operations, we can also use Awkward Array to perform averages along a specific axis. In our situation, ak.mean(lon, axis=1) perform the same mean operation per trajectory, see that the type is then reduced to subset_nb_drifters * float64.
*Note: The nan versions of those operations (ak.nanmean, ak.nanstd, ak.nanmax, etc.) is not yet available but is currently in development.
avg_lon = ak.mean(ds_ak.obs.lon, axis=1)
avg_lon
<Array [-53.5, -6.68, -74.5, ... -151, -143] type='500 * ?float64'>
Applying more complex operation can be easily implemented with simple for-loop, since the indexing is taken care by Awkward Array.
def periodogram_per_traj(uv):
dt = 1/24
d = []
for i in range(0, len(uv)):
d.append(dt*np.abs(np.fft.fft(uv[i]))**2)
return d
t0 = time.time()
ret = periodogram_per_traj(
ds_ak.obs.ve + 1j*ds_ak.obs.vn
)
benchmark_times[2,2] = time.time() - t0
Each element of the list ret now contains the fast Fourier transform of the complex velocity u + iv per trajectory:
ret[:5]
[<Array [48, 52.7, 4.64, ... 16, 58.1, 131] type='1095 * float64'>,
<Array [3.42e+05, 5.33e+04, ... 3.41e+03] type='19132 * float64'>,
<Array [4.62e+04, 6.59e+03, ... 3.11e+03] type='6631 * float64'>,
<Array [3.26e+05, 6.37e+04, ... 5.03e+04] type='17906 * float64'>,
<Array [1.08e+03, 774, 311, ... 540, 2.21e+03] type='5738 * float64'>]
Numba#
Awkward Array can be combined with Numba to accelerate calculations. Here, the goal is not to show the most efficient way of performing a calculation, but to present how simply adding the numba decorator @nb.njit to a function can improved the performance by order of magnitude(s).
Here we show an example that calculates the kinetic energy of a drifter trajectory from its velocity time series:
def kinetic_energy(u, v):
ke = np.zeros(len(u))
for i in range(0, len(u)):
ke[i] = np.sqrt(u[i]**2 + v[i]**2)
return ke
@nb.njit
def kinetic_energy_nb(u, v):
ke = np.zeros(len(u))
for i in range(0, len(u)):
ke[i] = np.sqrt(u[i]**2 + v[i]**2)
return ke
# select a random trajectory
i = 10
%%time
ke = kinetic_energy(ds_ak.obs.ve[i], ds_ak.obs.vn[i])
CPU times: user 160 ms, sys: 2.02 ms, total: 162 ms
Wall time: 164 ms
The function is compiled just in time when we execute the function for the first time, future executions are numba fast.
kinetic_energy_nb(ds_ak.obs.ve[i], ds_ak.obs.vn[i])
array([0.18460563, 0.19851099, 0.19387937, ..., 0.32900888, 0.32915556,
0.32936901])
%%timeit -r10 -n10
ke2 = kinetic_energy_nb(ds_ak.obs.ve[i], ds_ak.obs.vn[i])
705 µs ± 78.2 µs per loop (mean ± std. dev. of 10 runs, 10 loops each)
This results in more than a 500x sped up (!) for the same function simply by adding the @nb.njit decorators!
del ds_ak, ret, x_c, y_c
Discussion#
We presented a commonly used Lagrangian oceanographic dataset (the GDP dataset) which is currently available as individual NetCDF files (one of the means of distribution). Next, we presented an efficient data structure, the contiguous ragged array, to combine the variables from these individual NetCDF files. We then compared three different packages–xarray , Pandas, and Awkward Array—by performing typical Lagrangian workflow tasks.
Xarray
xarray is a well-known and established package used to perform operations on N-Dimensional datasets. It is natively integrated with Dask for parallel computing(not demonstrated in this Notebook), and allows users to lazy load data when the dataset is larger than the available memory of the computing environment.
However, it is not typically used with contiguous ragged array, so applying operation per trajectory requires to manually index through the dataset. One possibility is to set the size of the Dask chunks equals to the size of the trajectories, which allows for mapping operation in parallel per trajectory. However, the typical high number of chunks is less than ideal: it hinders performance since a simple operation like computing mean values across a dataset containing n trajectories requires to conduct n reading operation on the data file.
Pandas
Pandas is a predominantly used Python data analysis package. Since it is designed to perform operations on tabular data, it is required to form two DataFrame tables to represent the complete GDP dataset: one containing the variables with one value per trajectory (['traj']), and a second one containing the variables with one value per observations along the trajectory (['obs']). Unfortunately, this creates a separation the data and their metadata.
In this Notebook we used only a subset of the GDP dataset by randomly selecting only 500 trajectories. The data were therefore entirely contained within the active memory and the operations performed were fast and written in an elegant and intuitive fashion. However, if these operations are conducted on the entire dataset (17324 trajectories) we found the execution to be much slower than with xarray or Awkward Array which seem able to manage memory much more efficiently.
Awkward Array
Awkward Array is a novel library specifically designed to deal with ragged and heterogeneous data (in type or size). With the example of the heterogeneous GDP dataset, we find that this library simplifies considerably Lagrangian analysis. Furthermore, integration with Numba allows to speed up calculations without major redesign. We anticipate that these two advantages will lead to faster development of Lagrangian analysis functions as part of the CloudDrift project.
Initially developed for particle physics applications, Awkward Array is missing some useful features (e.g. html representation, import from NetCDF, and especially Dask integration) when compared to more mature libraries like xarray or Pandas. During the development of this Notebook, we had the opportunity to engage with the authors of this library, and we plan on collaborating to fill those gaps during the development the clouddrift libraries. We believe that this collaboration will elevate both projects, which are sponsored by the National Science Foundation during similar time frame (Awkward Array #2103945).
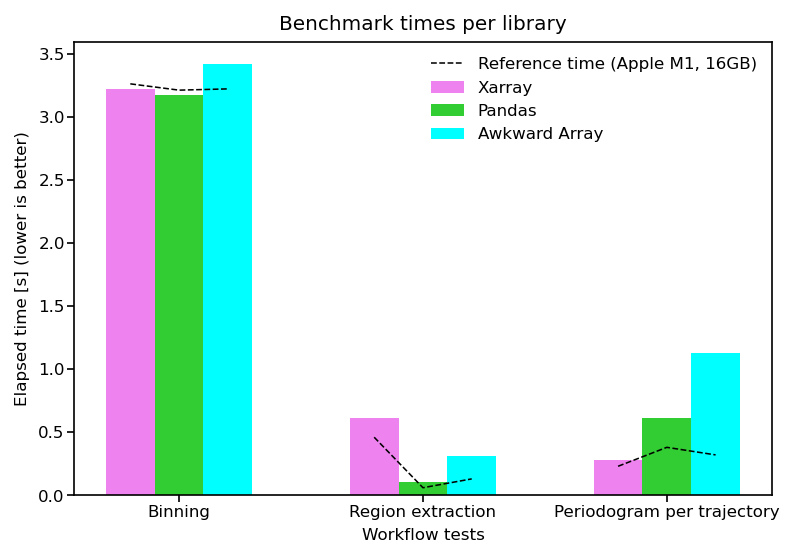
Benchmark speed#
The following figure presents the benchmark times obtain for the current execution. As a comparison, we include a reference time (black dashed line) obtained on a 2021 Mac Mini with a 8 cores Apple M1 ARM processor, 16GB of RAM, running macOS Monterey 12.3.
Note: user should expect slower time on a Binder or Google Colab environment. We understand that a lot of factors can influence the results of this benchmark, and it might not be the most accurate when running on a Cloud environment, or on a local cluster with multiple users. We still believe it gives an interesting view of the performance of the three selected libraries.
ref_times = np.array([[3.26, 0.46, 0.23], # [s]
[3.21, 0.06, 0.38],
[3.22, 0.13, 0.32]])
fig = plt.figure(dpi=150)
ax = fig.add_subplot(1,1,1)
index = np.arange(0,3)
bar_width = 0.2
t1 = plt.bar(index, benchmark_times[0,:], bar_width,
color='violet',
label='Xarray')
t2 = plt.bar(index + bar_width, benchmark_times[1,:], bar_width,
color='limegreen',
label='Pandas')
t3 = plt.bar(index + 2*bar_width, benchmark_times[2,:], bar_width,
color='aqua',
label='Awkward Array')
# add reference times
for i in range(0, len(index)):
x = np.array([index[i], index[i] + bar_width, index[i] + 2*bar_width])
ax.plot(x, ref_times[:,i], 'k', linewidth=0.75, linestyle='dashed', label='Reference time (Apple M1, 16GB)')
plt.xlabel('Workflow tests')
plt.ylabel('Elapsed time [s] (lower is better)')
plt.title('Benchmark times per library')
plt.xticks(index + bar_width, ('Binning', 'Region extraction', 'Periodogram per trajectory'))
handles, labels = plt.gca().get_legend_handles_labels()
by_label = dict(zip(labels, handles))
plt.legend(by_label.values(), by_label.keys(), frameon=False)
<matplotlib.legend.Legend at 0x17ff4c8e0>
Feedback#

If you ran this notebook on a local computer, please help us guide the development of CloudDrift by sharing one or all following points to the discussion thread on Github Discussion:
your comments on the ease of use of Xarray and/or Pandas and/or Awkward Arrays for Lagrangian data analysis;
the figure of your benchmark timings in the previous cell;
your system information generated by executing the next cell;
any other comments!
Thanks!
# summary of the system for log purpose
# inspired from https://stackoverflow.com/a/58420504/1558320
def getSystemInfo():
info={}
info['platform']=platform.system()
info['platform-release']=platform.release()
info['platform-version']=platform.version()
info['architecture']=platform.machine()
info['processor']=platform.processor()
try:
info['frequency']=psutil.cpu_freq(percpu=False)
except:
info['frequency']='N/A'
info['cores']=psutil.cpu_count(logical=False)
info['threads']=psutil.cpu_count(logical=True)
info['ram']=str(round(psutil.virtual_memory().total / (1024.0 **3)))+" GB"
return info
getSystemInfo()
{'platform': 'Darwin',
'platform-release': '21.4.0',
'platform-version': 'Darwin Kernel Version 21.4.0: Fri Mar 18 00:47:26 PDT 2022; root:xnu-8020.101.4~15/RELEASE_ARM64_T8101',
'architecture': 'arm64',
'processor': 'arm',
'frequency': 'N/A',
'cores': 8,
'threads': 8,
'ram': '16 GB'}
Future development#
We hope you will follow the development of CloudDrift, and encourage everyone to share this Notebook as well as submit feedback and comments to team members.
References#
Elipot et al. (2016), “A global surface drifter dataset at hourly resolution”, J. Geophys. Res. Oceans, 121, doi:10.1002/2016JC011716
Hansen, D. V., & Poulain, P. M. (1996). Quality control and interpolations of WOCE-TOGA drifter data. Journal of Atmospheric and Oceanic Technology, 13(4), 900-909, doi:10.1175/1520-0426(1996)013%3C0900:QCAIOW%3E2.0.CO;2
